Raman
Raman spectroscopy open source library of biomolecules set to boost AI-driven medical diagnostics
Nov 26 2025
Researchers at the Universitat Oberta de Catalunya and the Institute of Photonic Sciences have released an open Raman spectroscopy database that contains spectra for 140 key biomolecules, to support artificial intelligence models for biomolecule identification, disease diagnosis and biomedical research
Researchers at the Universitat Oberta de Catalunya (UOC), Barcelona, Spain, and the Institute of Photonic Sciences (ICFO), Castelldefels, Spain, have created an open Raman spectral database that is accessible to the scientific community and contains reference spectra for 140 major classes of biomolecules, including nucleic acids, proteins, lipids and carbohydrates.
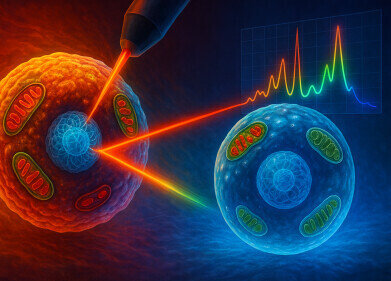
Raman spectroscopy is the vibrational spectroscopic technique that enables the analyse of chemical composition and molecular structure of materials through the interaction of light with matter. It relies on the phenomenon of Raman scattering, first described in 1928 by the Indian physicist Sir Chandrasekhara Venkata Raman, in which a small fraction of incident photons exchange energy with molecular vibrations and emerge with shifted wavelengths that act as highly specific molecular fingerprints. This work won him the 1930 Nobel Prize in Physics.
This study was led by data engineer and researcher Marcelo Terán, a doctoral candidate in the UOC’s Artificial Intelligence for Human Well-being (AIWELL) group, in collaboration with fellow AIWELL researchers Professors David Masip and David Merino, and ICFO scientists Dr. Pablo Loza-Álvarez and José Javier Ruiz. The project aimed to address one of the main bottlenecks that has limited the wider use of Raman spectroscopy in biomedicine: the lack of standardised, high-quality, open spectral libraries for biological molecules.
“One of the limitations of the potential of Raman spectroscopy in biomedical applications to date has been the lack of open spectral data for biomolecules. That is why we set out to create an accessible, standardised and useful library for the scientific community, which will act as the basis for future research and clinical applications,” said Terán, who is in the fourth year of his doctoral studies with the AIWELL group. By structuring the resource as an openly available reference library, the team has sought to lower barriers for laboratories that wish to incorporate Raman spectroscopy into workflows for biological analysis, without the need to generate all reference spectra from scratch.
In the project, the researchers implemented two search algorithms that they validated with measurements of pure biomolecules. The algorithms achieved 100% accuracy both in top ten identification of specific molecules – such as collagen – and in the correct assignment of the broader molecular class, such as protein, when they replicated the results of earlier benchmark studies. This level of performance suggests that the library can serve as a reliable backbone for computational tools that use Raman spectra to classify unknown biological samples at both molecular and functional levels.
“Raman spectroscopy can be used to analyse the chemical composition of samples in a non-invasive way, which is very valuable in the field of medicine.
“This database can facilitate the precise identification of biomolecules and, in the future, it will contribute to studying how their presence varies in biological processes such as cancer,” said Terán.
“The availability of high-quality biomedical data is essential for progress in the development of AI-based solutions. This need was the starting point for the research,” he added.
By linking robust physical measurements with curated digital resources, the work aligns spectroscopic practice with current trends in data-driven biomedical science.
To build the library, the researchers collected Raman spectral data for biomolecules from leading articles in the field and then developed an algorithm that used classical computer-vision techniques to extract numerical spectra automatically from published figures. Many Raman datasets exist only as plots embedded in journal articles rather than as downloadable numerical files, so an automated extraction approach has been necessary to reconstruct the underlying data in a reusable format.
One of the main challenges in the project was the limited amount of spectral data that authors had published in fully open-access, machine-readable form, which the team addressed through careful experimental validation of reconstructed spectra against laboratory measurements.
“Our work provides a tool that can help identify molecular composition based on its Raman spectrum in an objective, fast and standardised way. This identification is currently carried out by visual analysis of the main peaks in the spectra and is compared with the references in the literature.
“Our tool can streamline this process while providing a standard solution that reduces human bias during analysis,” said Terán.
By formalising the comparison step and embedding it in algorithms rather than subjective visual inspection, the library aims to support more reproducible analytical protocols in laboratories that use Raman spectroscopy for biological and clinical research.
Looking ahead, the researchers expect the scientific community to contribute further spectra to expand the database so that it evolves into a leading collaborative Raman spectral library for biomolecules.
“It is still unusual for scientific articles to share data openly, especially in the field of Raman spectroscopy. This lack of access to data limits biomedical research considerably.
“If AI is to be successfully applied, it needs large volumes of reliable and accessible data, and this is where open science projects play a key role,” said Terán.
The team hopes that journals, research groups and infrastructure providers will adopt policies that encourage systematic sharing of raw Raman spectra alongside published articles.
As the database grows, the authors anticipate that it will support the training of artificial intelligence models for the molecular analysis of biological samples. These models could help to distinguish subtle spectral differences that correspond to disease states, therapeutic responses or biochemical pathway activity. This, in turn, may create opportunities for novel applications in the diagnosis and monitoring of diseases, where Raman spectroscopy could operate as a label-free, non-destructive and potentially real-time analytical tool at the point of care or in specialised diagnostic laboratories.
For several years, the UOC has acted as a benchmark institution in the field of open science. It supports work of this kind through its Open Science Office, which promotes open data, open access and transparent research practices across disciplines. All the knowledge produced by the university can be found in open-access format in the O2 institutional repository, which already hosts articles, datasets, theses and teaching materials. The Raman spectral library now joins this ecosystem as a resource that aims to connect photonics, biomedicine and artificial intelligence under a shared commitment to openness and reproducibility.
For further reading please visit: 10.1016/j.chemolab.2025.105476
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh