-
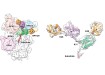
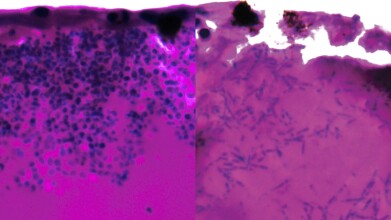 C.andida uris infecting fish larvae. On the left, replicating as a yeast, on the right, as a filament. Credit: University of Exeter
C.andida uris infecting fish larvae. On the left, replicating as a yeast, on the right, as a filament. Credit: University of Exeter
Research news
Genetic vulnerability in deadly Candida auris offers potential drug target
Jan 07 2026
Researchers at the University of Exeter have uncovered a genetic process in the emerging fungal pathogen Candida auris that could point to new treatment strategies for this deadly infection.
Candida auris has caused hospital outbreaks worldwide since its discovery in 2008, with a mortality rate of around 45% in critically ill patients. The fungus can resist all major classes of antifungal drugs, making infections difficult to treat and eradicate, particularly in intensive care units. In the UK, cases have steadily risen, highlighting its growing threat.
To investigate how the pathogen behaves during infection, the Exeter team pioneered a model using Arabian killifish larvae, which can survive at human body temperature. This allowed the researchers to study how C. auris genes are switched on during infection in a live host - something that had not previously been possible.
The study [1], published in Communications Biology, found that the fungus can form elongated filamentous structures, potentially to search for nutrients, and activates genes responsible for iron-scavenging. These findings suggest a potential “Achilles heel” that could be exploited with existing or novel drugs.
Co-senior author Dr Rhys Farrer, of Exeter’s MRC Centre for Medical Mycology, said: “Until now, we’ve had no idea what genes are active during infection of a living host. The fact that C. auris activates iron-scavenging genes gives clues to its origin and presents a possible target for treatment.”
NIHR Clinical Lecturer Hugh Gifford added: “While more research is needed to confirm this in humans, our findings could guide repurposing of drugs to stop C. auris from spreading in hospitals and affecting vulnerable patients.”
The research was supported by Wellcome, the Medical Research Council, and the National Center for Replacement, Reduction and Refinement (NC3Rs), which funded the killifish model as an alternative to traditional animal studies. Dr Katie Bates, NC3Rs Head of Research Funding, commented: “This study highlights how innovative alternative models can provide unprecedented insight into infections while reducing reliance on conventional animal experiments.”
This discovery represents a promising step toward understanding and targeting C. auris, a pathogen increasingly recognised as a global health threat.
More information online
- Xenosiderophore transporter gene expression and clade-specific filamentation in Candida auris killifish (Aphanius dispar) infection published in Communications Biology
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh