-
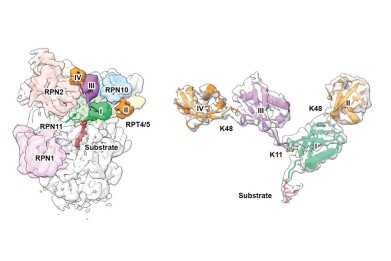 Cryo-EM structure of a K11/K48-branched ubiquitin chain bound to the 19S regulatory particle of the 26S human proteasome. (Left) The proximal ubiquitin (first ubiquitin attached to the substrate), K48-linked ubiquitin and K11-linked ubiquitin are colored green, orange, and magenta, respectively. The proteasomal subunits involved in ubiquitin binding are indicated. (Right) Expanded view of the K11/K48-branched tetra ubiquitin chain with the resolved cryo-EM map superimposed with the atomic model. Credit: National Taiwan University
Cryo-EM structure of a K11/K48-branched ubiquitin chain bound to the 19S regulatory particle of the 26S human proteasome. (Left) The proximal ubiquitin (first ubiquitin attached to the substrate), K48-linked ubiquitin and K11-linked ubiquitin are colored green, orange, and magenta, respectively. The proteasomal subunits involved in ubiquitin binding are indicated. (Right) Expanded view of the K11/K48-branched tetra ubiquitin chain with the resolved cryo-EM map superimposed with the atomic model. Credit: National Taiwan University
Research news
Atomic-level view reveals how the proteasome deciphers branched ubiquitin signals
Dec 10 2025
Researchers at National Taiwan University and Academia Sinica have, for the first time at atomic resolution, unveiled how the human proteasome recognises branched ubiquitin chains — a discovery that sheds new light on protein quality control and could have broad implications for understanding cellular regulation.
Protein degradation is central to maintaining cellular health, and the ubiquitin–proteasome system (UPS) serves as the cell’s primary mechanism for identifying and eliminating unwanted or damaged proteins. While homotypic K48-linked polyubiquitin chains have long been considered the canonical signal for degradation, how the proteasome reads these signals — especially more complex branched chains — has remained largely mysterious.
The team, led by Dr Shang-Te Danny Hsu, used an integrated approach combining cryo-electron microscopy, ubiquitin absolute quantification mass spectrometry (Ub-AQUA), and cross-linking mass spectrometry to capture the first high-resolution structure of the proteasome bound to a branched K11/K48-linked ubiquitin chain. Their findings [1] reveal a previously unrecognised K11-specific binding site on the proteasomal subunit RPN2. Together with other subunits, this forms a multivalent recognition groove that enhances substrate specificity and accelerates protein degradation.
“This integrated ‘structure + topology + absolute quantification’ strategy allowed us to resolve a question that has challenged the field for years,” said Professor Hsu. The study demonstrates that cells can rapidly eliminate key regulatory proteins, fine-tuning responses to stress and controlling protein homeostasis with remarkable precision.
Published in Nature Communications, the work represents a significant step forward in understanding how the proteasome deciphers complex ubiquitin signals and opens new avenues for research into diseases linked to protein degradation errors, including neurodegeneration and cancer.
More information online
- Structural basis of K11/K48-branched ubiquitin chain recognition by the human 26S proteasome published in Nature Communications
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh