-
-
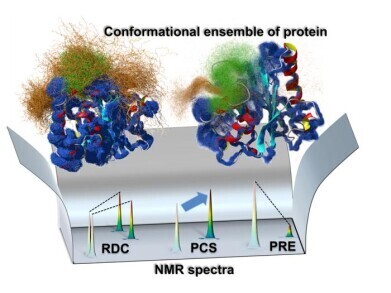 Ensemble map of protein structure. The team’s approach integrates different NMR data to create a map of how different parts of a molecular structure move. Credit: Tokyo Metropolitan University
Ensemble map of protein structure. The team’s approach integrates different NMR data to create a map of how different parts of a molecular structure move. Credit: Tokyo Metropolitan University
Microscopy & microtechniques
Tokyo team unveils novel NMR method to reveal enzyme movements in unprecedented detail
Oct 23 2025
Researchers in Japan have developed a Nuclear Magnetic Resonance spectroscopy technique that captures how enzymes move and interact during catalysis
Researchers at Tokyo Metropolitan University have developed a novel structure-determination method using Nuclear Magnetic Resonance (NMR) spectroscopy to show how different parts of complex molecular machinery – such as enzymes – move while they catalyse reactions.
Focusing on a specific enzyme in yeast, the team demonstrated how contrasts in atomic-scale motion influence biological function. The approach has provided an unprecedented means to explore the molecular mechanisms that underlie enzymatic activity and its links to disease.
Enzymes are indispensable to the function of all living organisms, including humans. Their detailed structures have been visualised through X-ray crystallography and cryo-electron microscopy, yet these methods typically capture static ‘snapshots’ of molecules, one at a time.
In their natural state, enzymes are in constant motion, shifting shape at the atomic level as they bind to other biomolecules and guide reactions through intricate, coordinated movements. These transient structural changes have long eluded direct observation due to the difficulty of capturing activity at the nanometre and millisecond scales.
The research team – led by Associate Professor Teppei Ikeya – combined several analytical approaches using NMR spectroscopy to capture the dynamic behaviour of enzymes in solution. Their method enabled the reconstruction of an accurate ‘ensemble structure’ the complete set of conformations a macromolecule can adopt, and the probability of each occurring.
To demonstrate the technique, the scientists examined yeast ubiquitin hydrolase 1 (YUH1), an enzyme responsible for recycling ubiquitin, a small protein that regulates numerous intracellular processes.
The human analogue of YUH1, known as ubiquitin C-terminal hydrolase (UCHL1), has been implicated in neurodegenerative disorders such as Parkinson’s disease and Alzheimer’s disease. By integrating multiple analytical datasets, the researchers created an ensemble map of YUH1 dynamics over the millisecond timescale.
They observed that two regions near the enzyme’s active site – the ‘crossover loop’ and the N-terminus, located at one end of the protein-binding structure – exhibited particularly large movements. The N-terminus appeared to swing in and out of the crossover loop, passing through a continuum of conformations before latching onto a substrate protein like a lasso.
Once bound, it acted as a ‘gating lid’ to hold the substrate in position for catalysis. This proposed mechanism was supported by experiments showing that mutant variants lacking a complete gating lid lost enzymatic activity.
The findings have revealed that the dynamic nature of enzymes is not merely incidental but integral to their catalytic precision. The researchers believe their NMR-based method can be extended to a wide range of biomolecular systems in near-native conditions, providing a transformative tool to investigate biological function and the molecular origins of disease.
For further reading please visit: 10.1021/jacs.5c06502
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh
.jpg)
-(2).jpg)