-
-
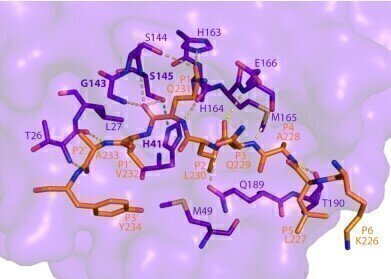 This image shows how SARS-CoV-2 Mpro recognizes and cuts NEMO based on the crystal structure determined using a powerful X-ray beam at SSRL Beam Line 12-2. (Credit : SLAC National Accelerator Laboratory)
This image shows how SARS-CoV-2 Mpro recognizes and cuts NEMO based on the crystal structure determined using a powerful X-ray beam at SSRL Beam Line 12-2. (Credit : SLAC National Accelerator Laboratory)
News
Critical Interaction reveals strength of SARS-CoV-2 Protein
Oct 06 2022
Scientists at the Department of Energy’s SLAC National Accelerator Laboratory recently witnessed the moment when a SARS-CoV-2 virus protein Mpro cut a protective protein known as NEMO, in an infected person. Without NEMO, an immune system is slower to respond to increasing viral loads or new infections.
Funnelling powerful x-rays from SLAC’s Stanford Synchrotron Radiation Lightsource (SSRL) onto crystallised samples of the protein complex, revealed how Mpro attacks NEMO at the molecular level as it dismantled NEMO’s primary function of helping our immune system communicate.
“We saw that the virus protein cuts through NEMO as easily as sharp scissors through thin paper,” said co-senior author Soichi Wakatsuki, professor at SLAC and Stanford. “Imagine the bad things that happen when good proteins in our bodies start getting cut into pieces.”
The images from SSRL showed the exact location of NEMO’s cut and provided the first structure of SARS-CoV-2 Mpro bound to a human protein.
“If you can block the sites where Mpro binds to NEMO, you can stop this cut from happening over and over,” SSRL lead scientist and co-author Irimpan Mathews said. “Stopping Mpro could slow down how fast the virus takes over a body. Solving the crystal structure revealed Mpro’s binding sites and was one of the first steps to stopping the protein.”
NEMO is a critical part of the human immune systems protective inflammatory response; when cut it helps the virus evade innate immune responses. A separate study by researchers at institutions in Germany found that the loss of NEMO by the action of Mpro could lead to damage in certain brain cells, causing neurological symptoms observed in COVID-19 patients, the researchers said.
“The crystal structures of NEMO and Mpro provide us with the targets to develop treatments that stop these cuts from happening,” SLAC scientist and co-first author Mikhail Ali Hameedi said. “Although current antiviral drugs can target Mpro, seeing the molecular details of how Mpro attacks NEMO will help us develop new treatments in the future as Mpro mutates.”
To predict how well Mpro binds to NEMO, researchers used the Summit supercomputer at the Oak Ridge Leadership Computing Facility, combining molecular dynamics simulations with five machine learning models. Applying quantum chemistry, they found that Mpro likely has the highest binding affinity in SARS-CoV-2 compared to the other primary coronaviruses.
“With a set of computational approaches, we were able to predict the strongest binding spots between NEMO and Mpro,” co-first author and ORNL scientist Erica Prates said. “We think that a high binding affinity at these hot spots helps explain the high fitness of the virus in humans.”
Moving forward, the biomedical industry could use the study to help build better inhibitor drugs and understand how other proteins could be affected by Mpro, Wakatsuki said.
“NEMO is only the tip of the iceberg,” he added.. “We can now study what happens when many other proteins in the body are cleaved by Mpro during infection.”
More information online
Citation: M. A. Hameedi et al., Nature Communications, 8 September 2022 (10.1038/s41467-022-32922-9)
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh