-
-
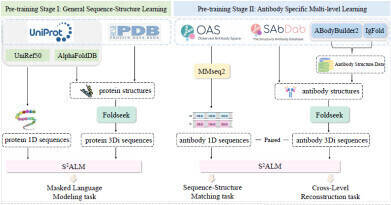 S2ALM’s two-stage framework: Stage I learns sequence–structure patterns from protein data, while Stage II applies antibody-specific training with Sequence–Structure Matching and Cross-Level Reconstruction. Credit: Copyright © 2025 Mingze Yin et al
S2ALM’s two-stage framework: Stage I learns sequence–structure patterns from protein data, while Stage II applies antibody-specific training with Sequence–Structure Matching and Cross-Level Reconstruction. Credit: Copyright © 2025 Mingze Yin et al
Research news
Chinese researchers develop AI model to design antibodies by combining sequence and structural data
Aug 28 2025
Researchers have developed a novel artificial intelligence model – S2ALM – that integrates sequence and structural data to design antibodies with greater accuracy, paving the way for faster and more cost-effective immune-based therapies
Antibodies – also known as immunoglobulins – are proteins produced by the body’s immune system to fight pathogens. Each antibody has a unique structure that enables it to bind to its target with high specificity. Owing to their precision and the relatively low risk of side effects, antibodies have been widely investigated as the basis for therapeutic drugs.
Traditionally, scientists have relied on the laborious wet laboratory methodologies to study and design antibodies. However, computational approaches now offer faster and more precise alternatives. In a major step towards the use of artificial intelligence (AI) in this field, a team of researchers in China has developed a novel model called the Sequence–Structure multi-level pre-trained Antibody Language Model (S2ALM). The system has been designed to analyse, predict and design antibodies by incorporating structural as well as sequence-based information.
The study was led by Professor Tingjun Hou and Professor Chang-Yu Hsieh from the College of Pharmaceutical Sciences, Zhejiang University, in collaboration with Assistant Professor Jintai Chen from AI Thrust, Information Hub, Hong Kong University of Science and Technology (Guangzhou), and Professor Jian Wu from the Zhejiang Key Laboratory of Medical Imaging Artificial Intelligence.
“The molecular basis of any antibody protein lies in its amino acid sequence,” said Professor Hou.
“The sequence decides its three-dimensional structure and the structure decides its biological function,” he added.
While most existing AI models have focused only on amino acid sequences, S2ALM has been the first to integrate both sequence and structure, thereby providing a more complete understanding of antibody function.
To train the system, the team compiled a dataset of more than 75 million antibody and protein sequences together with 11.7 million three-dimensional structures, including both experimentally determined and computationally predicted forms.
Two novel learning strategies were introduced into a hierarchical pre-training framework. The first, known as Sequence–Structure Matching, enabled the model to connect sequence data with structural features. The second, Cross-Level Reconstruction, allowed the model to predict missing information by combining sequence and structural clues.
The combination proved highly effective. S2ALM surpassed leading models in a range of tasks critical to antibody research and drug discovery. These included prediction of antigen-binding capacity, analysis of B cell maturation, identification of antibody paratopes, estimation of antigen–antibody binding affinity, and the design of novel antibody sequences.
One of the most striking outcomes has been the model’s ability to generate entirely novel antibody candidates with potential to target pathogens such as SARS-CoV-2 – the COVID-19 pandemic virus, Ebola and Influenza B viruses.
Structural predictions have indicated that these AI–designed antibodies can form stable, functional three-dimensional shapes suitable for therapeutic use.
“The success of S2ALM is three-fold; firstly, it learns from comprehensive antibody data, secondly, its learning approach combines structural and biological features, and thirdly, it has exceeded state-of-the-art performance in multiple tasks, including the design of novel antibodies,” said Professor Wu.
The development of S2ALM represents a milestone in antibody research by reducing reliance on the trial-and-error approach to experimentation. It has the potential to accelerate the creation of next-generation therapies, offering a faster, more reliable and cost-effective routes to bring immune-based treatments to the clinic.
For further reading please visit: 10.34133/research.0721
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh