Research news
Nipah's polymerase structure offers new target for antiviral drug development
Jun 13 2025
The molecular structure of the Nipah virus (NiV) polymerase complex has been resolved by a team of researchers from Guangzhou Medical University, China, providing key insights into how the virus replicates and offering a potential pathway for the development of broad-spectrum antiviral drugs.
NiV is a highly pathogenic, zoonotic virus that causes severe disease in humans, with high mortality rates and no approved antiviral treatments. Understanding the virus’s replication machinery is considered critical to developing targeted therapeutics that could prevent future outbreaks.
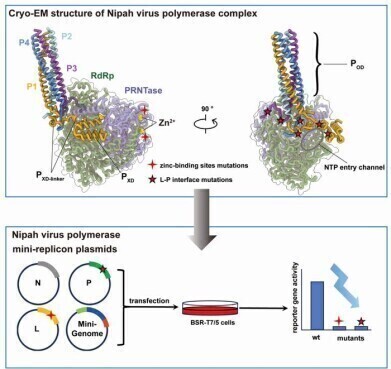
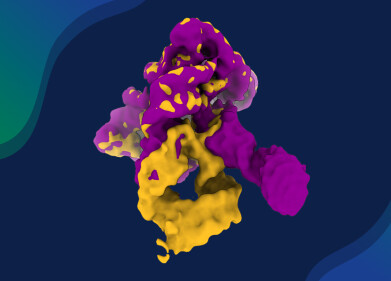
In a study published in the journal Protein & Cell earlier this year, researchers reported the cryo-electron microscopy (cryo-EM) structures of the viral RNA-dependent RNA polymerase (L protein) in complex with its phosphoprotein cofactor (P protein). Two structural states of the L–P complex were resolved: one featuring the full-length L protein, the other a truncated form. Both revealed defined RNA polymerase (RdRp) and capping enzyme (PRNTase) domains within the L protein, while the C-terminal domains remained unresolved due to structural flexibility.
The study identified two evolutionarily conserved zinc-binding motifs within the PRNTase domain that were essential for enzymatic activity. Disruption of these sites through alanine substitution completely abolished polymerase function. These motifs are conserved across most members of the Mononegavirales order, though absent in pneumoviruses.
The P protein was found to form a tetrameric bundle that interacted with the L protein through multiple hydrophobic, hydrogen bonding and salt bridge interactions. The XD domain of one P subunit anchored stably to the RdRp surface, while the linker regions of other subunits extended across the nucleotide entry channel, suggesting a possible regulatory role in RNA template accessibility.
Mutagenesis experiments targeting these interaction sites significantly reduced polymerase activity and compromised L protein stability, demonstrating their functional importance. Structural comparisons across other Mononegavirales, including Newcastle disease virus and Ebola virus, revealed a conserved tyrosine residue (Y732 in NiV) that stabilises the P protein’s anchoring to the L protein. This residue appears to play a pivotal role in complex formation across different viruses in the order.
The study marks a significant advance in the structural biology of henipaviruses. By elucidating the architecture of the viral polymerase and its interaction with regulatory cofactors, the findings lay the groundwork for rational antiviral drug design. Specifically, targeting the conserved interaction surfaces and catalytic sites identified in the study could facilitate the development of inhibitors with broad-spectrum activity against related viruses.
The authors concluded that their structural analysis not only enhances the mechanistic understanding of NiV replication but also provides a robust foundation for antiviral discovery. These insights are particularly timely given the continuing public health threat posed by emerging henipavirus outbreaks.
For further reading please visit: 10.1093/procel/pwaf014
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh