-
 Researchers from Japan have now identified RABGGTB as a novel autism-linked gene during DNA methylation mapping in the dorsal raphe nuclei of the brain. Credit: Hideo Matsuzaki from the University of Fukui, Japan
Researchers from Japan have now identified RABGGTB as a novel autism-linked gene during DNA methylation mapping in the dorsal raphe nuclei of the brain. Credit: Hideo Matsuzaki from the University of Fukui, Japan
Research news
Gene hunt using DNA methylation suggest RABGGTB as autism biomarker candidate
May 22 2025
Changes to key brain regions in analysis of ASD and non-ASD patients points to discovery of biomarker for autism spectrum disorder
Autism spectrum disorder (ASD) is a lifelong neurodevelopmental condition characterised by differences in social communication and behaviour, as well as unique patterns of interests and sensory experiences. Affecting brain development, recent studies have suggested that epigenetic processes and factors in the environment can alter DNA methylation.
A team in Japan, led by Professor Hideo Matsuzaki, Research Center for Child Mental Development, University of Fukui, with Dr. Keiko Iwata, School of Pharmaceutical Sciences, Wakayama Medical University, conducted an exploratory epigenetic study to profile postmortem brain samples of individuals with and without ASD.
Immune activation and stress hormone exposure alter neuronal activity in the dorsal raphe (DR) – which are a cluster of neurons located in the brainstem and the largest source of serotonin in the brain – and has been shown to contribute to the development of ASD.
Epigenetic profiling analyses chemical modifications on DNA that affect the regulation of genes. DNA methylation profiles of the DR in the context of ASD have not previously been investigated. DNA methylation is an epigenetic modification where a methyl group is added to DNA – which can regulate gene expression without changing the underlying genetic sequence – and often acting to silence a gene.
Using the Illumina Infinium HumanMethylation450 BeadChip array for DNA methylation profiling and qRT-PCR for gene expression analysis, the team undertook a genome-wide DNA methylation analysis focused on the DR nucleus. In addition, the researchers validated site-specific methylation using electron microscopy-amplicon sequencing.
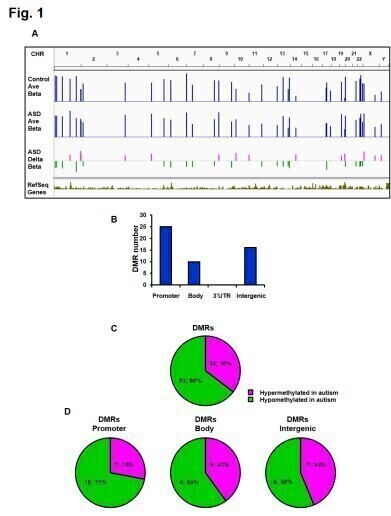
The study revealed extensive changes to DNA methylation in the DR genomic regions. Genes such as OR2C3 (olfactory receptor gene) and HTR2C (serotonin receptor gene) showed hypermethylation – increase in DNA methylation activity – in samples from ASD brains.
This identification of DNA methylation in these genes could be linked to differences in sensory processing and serotonin disruption in ASD patients. Further, hypomethylation was observed in the RABGGTB promoter region – a gene related to autophagy and synaptic function which corresponded to its heightened expression.
“RABGGTB is an exciting discovery. It is absent from the current SFARI gene database (a major scientific resource focusing on genes associated with ASD), making it a truly novel candidate for autism,” said Professor Matsuzaki, elaborated on the team’s findings.
“Studying this gene could open new doors to understanding ASD, and perhaps even lead to a future diagnostic biomarker,” he said.
Further study will be needed to combine DNA methylation profiling with transcriptome analysis to link DNA methylation with RNA expression.
“Our study provides valuable insights into the molecular landscape of ASD, opening avenues for future diagnosis and therapeutic breakthroughs for autism,” added Matsuzaki.
For further reading please visit: 10.1111/pcn.13830
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh