-
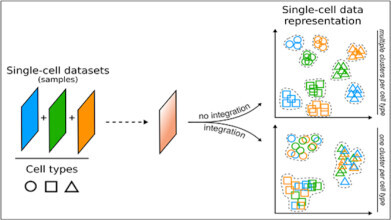 Illustration highlighting the difference between analysing single-cell data without or with integration. Credit: University of Turku
Illustration highlighting the difference between analysing single-cell data without or with integration. Credit: University of Turku -
 L-R: António Sousa and Sini Junttila. Credit: University of Turku
L-R: António Sousa and Sini Junttila. Credit: University of Turku
Research news
Machine learning untangles complex single-cell data
Nov 24 2025
Scientists at Turku Bioscience Centre, University of Turku, have developed a machine-learning method to make sense of one of single-cell analysis’ biggest challenges: comparing complex, uneven datasets across samples.
Modern single-cell technologies capture thousands of molecular measurements from individual cells, revealing extraordinary cellular diversity. But when researchers analyse multiple samples – each with different proportions of cell types, or cell types present in one sample but not another – existing computational methods often falter. Distinct cell populations can be mistakenly merged, masking biological differences.
The Turku team has tackled this using a new algorithm, Coralysis, developed in Professor Laura Elo’s Computational Biomedicine group. Instead of forcing datasets together in one step, Coralysis uses a staged integration approach inspired by assembling a jigsaw puzzle: cells are grouped from simple to increasingly detailed features, progressively refining cell identities with each clustering round.
The result is a tool that can reliably integrate imbalanced datasets and detect subtle shifts in cellular state that conventional methods frequently overlook.
“We wanted a method that uncovers these hidden patterns without collapsing rare or uneven cell types,” said Associate Professor Sini Junttila, who co-supervised the work.
Coralysis is available as open-source software, and its machine-learning engine can be trained to predict cell identities in entirely new datasets – with built-in confidence estimates. This avoids laborious manual annotation and improves reproducibility across studies.
Lead developer António Sousa described the goal: “Like assembling a puzzle, we start with simple features and build up. Coralysis integrates cells in the same stepwise way.”
Professor Elo added that openly releasing the tool will help accelerate discoveries across the field: “Coralysis gives researchers a new way to navigate cellular complexity.”
The study [1] by Elo’s research group has been published in the scientific journal Nucleic Acids Research.
More information online
- Coralysis enables sensitive identification of imbalanced cell types and states in single-cell data via multi-level integration published in Nucleic Acids Research
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh