-
-with-research-lead-Dr-Sandro-Ataide-and-first-author-Rezwan-Siddiquee-(right)---C.-Photo-Fiona-Wolf-USYD.jpg) Caitlin McCormack (left) with research lead Dr Sandro Ataide and first author Rezwan Siddiquee (right) - C. Photo Fiona Wolf-USYD
Caitlin McCormack (left) with research lead Dr Sandro Ataide and first author Rezwan Siddiquee (right) - C. Photo Fiona Wolf-USYD -
 Dr Sandro Ataide - office - University of Sydney. C -Photo by Fiona Wolf-USYD
Dr Sandro Ataide - office - University of Sydney. C -Photo by Fiona Wolf-USYD -
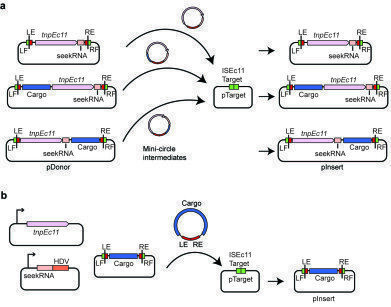 a:Schematic of transposition assay for ISEc11 carrying cargo. The pDonor plasmids include ISEc11 or ISEc11 with the cargo DNA of interest (blue box labelled cargo) inserted upstream (to the left) or downstream (to the right) of the tnp gene (large pink box). pTarget included the ISEc11 target (green box). The seekRNA (small pink box and the DNA sequence from the IS ends (red boxes) and flanks including the target (green boxes) are also indicated. The mini-circle intermediate and predicted transposition products are also shown. b: Schematic of mini IS transposition assay showing the three compatible plasmids present. The mini IS contains the cargo DNA flanked by DNA sequence from the left IS end (LE) and from the right end (RE) and is flanked by target sequences. The tnpEc11 (pink box) and the seekRNA followed by a HDV ribozyme (light and dark orange boxes) are in separate plasmids. Other colours are as in a. Movement of the mini IS into a fourth plasmid, pTarget containing the ISEc11 target. *
a:Schematic of transposition assay for ISEc11 carrying cargo. The pDonor plasmids include ISEc11 or ISEc11 with the cargo DNA of interest (blue box labelled cargo) inserted upstream (to the left) or downstream (to the right) of the tnp gene (large pink box). pTarget included the ISEc11 target (green box). The seekRNA (small pink box and the DNA sequence from the IS ends (red boxes) and flanks including the target (green boxes) are also indicated. The mini-circle intermediate and predicted transposition products are also shown. b: Schematic of mini IS transposition assay showing the three compatible plasmids present. The mini IS contains the cargo DNA flanked by DNA sequence from the left IS end (LE) and from the right end (RE) and is flanked by target sequences. The tnpEc11 (pink box) and the seekRNA followed by a HDV ribozyme (light and dark orange boxes) are in separate plasmids. Other colours are as in a. Movement of the mini IS into a fourth plasmid, pTarget containing the ISEc11 target. *
Research News
seekRNA: Advancing precision gene editing beyond CRISPR
Jul 11 2024
Scientists at the University of Sydney have unveiled a gene-editing tool named seekRNA, which offers superior accuracy and flexibility compared to the widely-used CRISPR technology. This new tool, developed by Dr Sandro Ataide and his team at the School of Life and Environmental Sciences, utilises a programmable RNA strand to precisely target and relocate genetic sequences, thereby simplifying the editing process and minimising errors. Their research has been published in Nature Communications [1].
Dr Ataide highlights the transformative potential of seekRNA, emphasising its ability to deliver precise and flexible target selection, thus heralding a new era in genetic engineering. Unlike CRISPR, which requires additional components to function as a ‘cut-and-paste’ tool, seekRNA operates as a standalone system, enhancing accuracy and broadening the range of DNA sequences it can modify.
Traditional CRISPR technology involves creating breaks in the DNA double helix and relies on other proteins or the cell’s DNA repair machinery to insert new sequences, often introducing errors. In contrast, seekRNA can cleanly cleave the target site and insert new DNA sequences without needing additional proteins, resulting in higher accuracy and fewer errors.
Since its inception over a decade ago, gene editing, particularly through CRISPR, has revolutionised research and applications across various fields. It has improved disease resistance in agriculture, expedited human disease detection, and contributed to advanced cancer treatments like CAR T-cell therapy. Despite these advances, Dr Ataide notes that the field is still in its early stages, and seekRNA's development aims to further propel health, agricultural, and biotechnological innovations.
SeekRNA is derived from the IS1111 and IS110 families of naturally occurring insertion sequences found in bacteria and archaea. These sequences exhibit high target specificity, which seekRNA leverages to achieve its impressive accuracy. The tool can be modified to target any genomic sequence and insert new DNA with precise orientation. Laboratory tests have shown promising results in bacteria, and future research will focus on adapting the technology for complex eukaryotic cells found in humans.
One of seekRNA's advantages is its compact size, consisting of a small protein of 350 amino acids and an RNA strand of 70 to 100 nucleotides. This allows it to be delivered efficiently using biological nanoscale delivery vehicles like vesicles or lipid nanoparticles. Additionally, seekRNA can independently insert DNA sequences in desired locations, a capability that many current editing tools lack.
University of Sydney research associate Rezwan Siddiquee, lead author of the study, points out that current CRISPR technology has limitations on the size of genetic sequences it can introduce, which restricts its applications. In contrast, seekRNA’s small and efficient system can move genetic cargo more effectively.
Globally, research teams are exploring the gene-editing potential of the IS1111 and IS110 families, but Dr Ataide’s team is at the forefront, having demonstrated results for a member of the IS110 family and advancing the technique with the shorter seekRNA. The University of Sydney’s pioneering work continues to drive innovation in gene editing, promising significant advancements in the field.
More information online
1. Siddiquee, R. et. al. ‘A programmable seekRNA guide target selection by IS1111 and IS110 type insertion sequences’. (Nature Communications) DOI: 10.1038/s41467-024-49474-9
Digital Edition
Lab Asia 31.6 Dec 2024
December 2024
Chromatography Articles - Sustainable chromatography: Embracing software for greener methods Mass Spectrometry & Spectroscopy Articles - Solving industry challenges for phosphorus containi...
View all digital editions
Events
Jan 22 2025 Tokyo, Japan
Jan 22 2025 Birmingham, UK
Jan 25 2025 San Diego, CA, USA
Jan 27 2025 Dubai, UAE
Jan 29 2025 Tokyo, Japan