Mass spectrometry & spectroscopy
In recent decades, genomics has powered much of our progress in precision medicine. The ability to map millions of genetic variations and sequence whole genomes has enabled researchers to identify disease-associated variants, stratify risk, and advance our understanding of human biology. Building on this foundation, deeper biological insights are needed to expand the therapeutic development and clinical opportunities that can offer new approaches to treat disease and illness. Our genes may provide the blueprint of inherited potential, but they don’t tell the full story of what’s happening in the body right now.
Enter proteins. Proteins, the functional products of our genes, serve as real-time indicators of biological processes, reflecting both genetic and environmental influences. Unlike the static nature of DNA, proteins fluctuate with disease progression, therapy response, aging and lifestyle factors. As a result, proteomics (the large-scale study of proteins) offers the missing layer of insight needed to fulfil the promise of precision medicine.
We are now entering a new frontier in translational research for human health, one defined not only by sequencing the genome, but by quantifying the proteome and how it varies, at scale. And with technological advances across analytical and diagnostic instruments enabling the measurement of thousands of proteins across hundreds of thousands of samples, proteomics is poised to transform how we understand, diagnose and treat disease.
Proteomics bridges the gap between genotype and phenotype by providing real-time snapshots of biological function. While genomics reveals potential risk inherited at birth, proteomics reflects what is happening at a given point in time, which makes it more suitable for disease monitoring, therapeutic targeting, and outcome prediction.
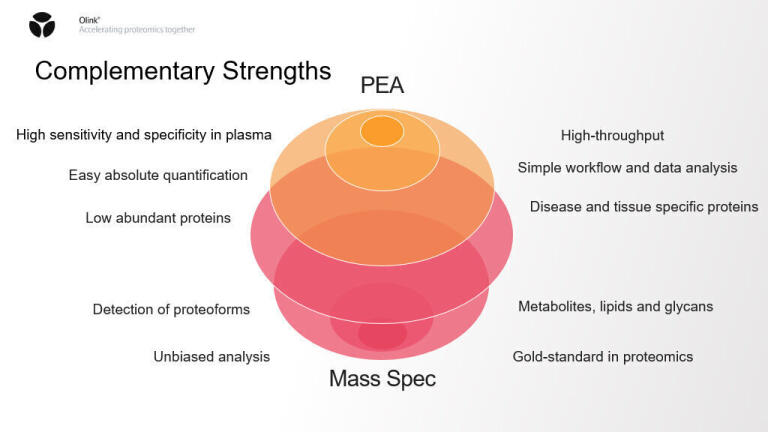
The journey from discovery to clinical translation demands both breadth and precision. To meet this challenge, researchers rely primarily on two complementary proteomics modalities: mass spectrometry and affinity-based assays, like the proximity extension assay (PEA) used by Olink® and others.
Mass spectrometry, powered by cutting-edge instruments like the Thermo Scientific™ Orbitrap™ Astral™ Zoom and Stellar™ mass spectrometers, offers unparalleled depth and resolution [1]. Mass spectrometry-based proteomics excels in characterising proteoforms (specific molecular forms of a protein), post-translational modifications at peptide resolution, with particular versatility in tissues including single cell proteomes, with a throughput of up to 500 samples per day per instrument line.
Affinity-based assays like Olink’s PEA [2] are designed for population-scale throughput, minimal sample requirements and an emphasis on low abundant proteins typically found in plasma. With as little as 2 microlitres, Olink can analyse over 5,400 proteins in over 2,000 samples per day per sample line, complementing the impact of MS-based analysis. This makes PEA particularly suited for large cohort studies, where processing volume and sample conservation are critical.
Each with their own strengths, these technologies are increasingly used in tandem. Mass spectrometry provides granular, peptide-level insights, while affinity-based methods offer high-throughput screening across broad protein panels. Together, these technologies form the foundation of a multi-omics strategy that integrates genomic, proteomic, and clinical data to fuel both discovery and clinical utility.
The power of proteomics is best illustrated through its applications. Across the globe, large-scale studies are harnessing proteomic data to illuminate disease biology and inform therapeutic development.
The UK Biobank Pharma Proteomics Project [3[, arguably the most ambitious of its kind, is a landmark example. In collaboration with Olink and funded by 14 pharma partners, the project will generate proteomic data on 600,000 individuals, most with corresponding genomic and clinical data. The pilot phase alone, comprising 54,000 samples and ~3,000 proteins, has already yielded over 200 peer-reviewed publications, been referenced in over 750 others, been accessed over 175,000 times and enabled predictive modelling across over 200 diseases.
In one notable example, researchers built a ‘protein clock’ using UK Biobank data combined with other data from biobanks in Finland and China [4]. This composite biomarker outperformed methylation clocks in predicting biological age, highlighting the value of measuring proteins as real-time indicators of physiological state. These clocks could potentially be used to identify individuals who are aging faster than expected, allowing for earlier interventions and more personalised treatments.
Elsewhere, in a landmark study [5] from Oxford University and funded by Cancer Research UK, researchers applied proteomics to identify early biomarkers for cancer through use of over 44,000 individuals from the UK Biobank representing 19 blood and solid tumour cancers. They revealed predictive protein signatures a median of 12 years before diagnosis.
Also focused on cancer, a team out of KTH Royal institute of Technology [6] in Sweden led an initiative to build a pan-cancer proteomic atlas and pinpointed high precision ovarian cancer biomarkers in plasma, signalling progress toward less-invasive diagnostics in a disease where earlier detection has critical implications for survival.
Beyond emerging research, proteomics holds great promise for clinicians and patients selecting the right treatment options and avoiding unnecessary interventions. For instance, Octave Biosciences has developed an 18-protein panel for multiple sclerosis disease activity monitoring [7]. This panel has been validated clinically and now used in leading MS Centers of Excellence, academic institutions and independent neurology practices across all 50 states in the United States as part of a disease activity dashboard to support treatment decisions.
Similarly, research into protein analysis offers a promising pathway to personalised medicine for individuals with inflammatory bowel disease, including ulcerative colitis. A study out of the University of Edinburgh revealed that a more nuanced approach to understanding molecular differences of this disease would allow physicians to better predict which individuals would respond to biological therapeutics [8]. Work being done through the Crohn’s and Colitis Foundation has extended this to paediatric cohorts, building a machine learning informed multi-omic signature to identify children with Crohn’s disease who were at higher risk for complications and predict treatment response.
Projects like these demonstrate proteomics’ growing role not only in discovery science, but also in building disease-specific tools that are poised to fuel new innovation that will enter the clinic. Proteomic science is truly moving beyond early-stage discovery with promise to detect disease earlier, stratify patients more precisely, and accelerate therapy development.
Affinity-based platforms like Olink’s provide critical advantages in sensitivity and scalability. This technology excels at detecting low-abundance proteins, including those implicated in early-stage disease, and it can do so with high specificity using minimal sample volumes and at very large sample throughput. This is a key reason why large biobank projects increasingly turn to this modality.
Meanwhile, mass spectrometry adds value by drilling down to the peptide level, enabling scientists to distinguish among protein variants and modifications that may alter function, which is a capability not typically accessible with affinity-based methods.
Together, these complementary approaches enhance our ability to detect dynamic biological changes, whether tracking disease progression or measuring therapeutic response.
One of the most persuasive reasons to invest in proteomics is its demonstrated ability to outperform genomics in predicting disease outcomes. In a landmark study by Gad et al., researchers used UK Biobank data to compare polygenic risk scores with protein-based models across 23 diseases [9].
For type 2 diabetes, protein scores achieved 89% predictive accuracy - more than 20 points higher than genomic scores alone. Similar trends were observed for other complex diseases, including cancer and cardiovascular conditions. These improvements reflect the fact that proteins capture downstream effects of both genes and environment, offering a more immediate and relevant view of disease biology.
Furthermore, proteomic data enhances clinical trial success. By identifying causal proteins through Mendelian randomisation, as demonstrated in the SCALLOP consortium, researchers can pinpoint druggable targets with higher precision. In one analysis, 25 potentially causal therapeutic targets were identified, 56% of which were immediately corroborated as known drug mechanisms [10]. These results emphasise the translational power of these datasets.
Building an atlas of integrated genomics, proteomics, and clinical outcomes serves as a critical resource for both pharma and academic partners, improving both trial design and therapeutic development.
As proteomics matures, its future lies in deeper integration, finer resolution and broader accessibility. Emerging fields such as spatial proteomics and single-cell proteomics promise to map protein expression with cellular and tissue-level granularity, which will open new frontiers in understanding disease microenvironments.
Meanwhile, innovations in data sharing and standardisation are enabling a “measure once, query often” model. This paradigm is especially powerful in global consortia, where datasets can be leveraged across diverse research questions, allowing maximum impact while minimising redundancy.
One particularly promising area is the study of extracellular vesicles - tiny protein-laden packets circulating in plasma. Even after freezing, these vesicles preserve their content, and affinity-based assays can detect them in minuscule sample volumes. As tools evolve to profile the vesicles in detail, we may uncover new biomarkers and signaling mechanisms central to disease dynamics.
Finally, artificial intelligence stands to accelerate proteomics’ clinical relevance. But as leaders in autonomous systems have learned, AI is only as good as the data it ingests. In healthcare, that means prioritising specificity and accuracy, which are two hallmarks of high-quality proteomic datasets.
By fuelling AI models with the best available biological data, we can build smarter diagnostics, predict patient trajectories and personalise therapy more effectively than ever before. As workflows become more automated and accessible, proteomics is poised to become as foundational as genomics in research and diagnostics. Spatial proteomics, single-cell analysis, and standardised multi-site data integration will further extend its utility.
Yet the core enabler remains the same: rigorous, specific, scalable, and reproducible protein measurement that translates molecular data into real-world outcomes.
From large-scale biobank studies to clinical implementation in diseases like multiple sclerosis, ovarian cancer and ulcerative colitis, proteomics is reshaping our approach to healthcare. It complements genomic insight with real-time biological data, deepens our understanding of disease mechanisms, and empowers predictive, personalised healthcare. The future will be shaped by the ability to drive a broad set of customisable tools tailored to support decision-making for meeting the needs of patients. In this new era, proteins are more than just the workhorses of biology. They are the next great biomarkers in frontier of precision medicine.