-
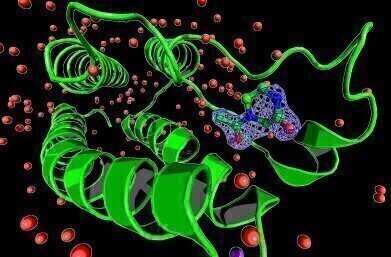 Electron density for a ligand bound to bromodomain BRD1 obtained using PanDDA method.
Electron density for a ligand bound to bromodomain BRD1 obtained using PanDDA method.
News
Hidden 3D Data Unveiled by PanDDA
Jun 29 2017
A new method to extract previously hidden information from the X-ray diffraction data that are measured when resolving the three-dimensional (3D) atomic structures of proteins and other biological molecules has been developed by scientists utilising Diamond Light Source.
When studying the atomic detail of how compounds bind to their target proteins, for example in rational drug design, scientists compare X ray data measured in both presence and absence of the compound. However, with existing analysis algorithms, this difference signal can often be swamped by noise from experiment artefacts, making it very unreliable to interpret the observed signal.
The new Pan-Dataset Density Analysis (PanDDA) method* extracts the picture of the bound compound by first identifying the source of the noise and removing it from the data. It exploits Diamond’s ability to repeat dozens to hundreds of measurements quickly, which are then characterised for differences between them, indicating the presence of bound compound, after which a noise correction is applied in 3D.
“The problem of identifying binding events in crystallographic datasets can feel like looking for a needle in a haystack,” explains Dr Nicholas Pearce, lead author on the paper which comes from his PhD project at the University of Oxford in the Systems Approaches for Biomedical Science (SABS) Centre for Doctoral Training, where he was jointly funded by UCB Pharma and Diamond. “In the case of the data we were analysing, it was even worse, because we had hundreds of haystacks and didn’t know which of them contained needles.” Dr Pearce is now based in the Crystal & Structural Chemistry Group at Universiteit Utrecht.
The researchers were able to use to their advantage the fact that most of the measurements were from ‘empty’ crystals that didn’t contain a bound ligand, allowing them to characterise the unbound form and simply looking for datasets that were different.
“Often in crystallography you can miss ‘weak’ bound forms, because each measurement is a superposition of the bound and unbound forms,” continues Dr Pearce. “This is akin to multiple sheets of tracing paper, each with one of at least two images, all overlaid on top of each other.”
“When trying to identify the image on only one of the ‘sheets’, it gets confused by what shows through from all the other sheets, so the image becomes susceptible to interpretation errors,” Dr Pearce adds. “To overcome this, we developed a method to extract the right set of ‘sheets’ from the superposition; once we’d done that, interpreting the bound form becomes much easier and enables us to confidently interpret the data and build models of the interesting states in the data.”
“The basic idea is conceptually very simple, namely treating the confusing superposition as a background correction problem,” explains Professor Frank von Delft, who is jointly Principal Investigator of the Protein Crystallography group in the Structural Genomics Consortium (SGC) at the University of Oxford and Principal Beamline Scientist of the I04-1 beamline at Diamond. “However, an accurate estimate of the background is crucial and in practice this was unthinkable until the advent of the new robotic technology offered by Diamond, which makes it routine to make such large numbers of measurements.”
“UCB is delighted to have been working closely with Diamond on the development of PanDDA and its application to crystallographic fragment screening,” comments Dr Neil Weir, Senior Vice President of Discovery at UCB Pharma. “As a direct result, we have been able to identify fragments, which were otherwise not distinguishable from background, bound to a key protein-protein interaction drug target.”
*The results are published in Nature Communications
Digital Edition
Lab Asia Dec 2025
December 2025
Chromatography Articles- Cutting-edge sample preparation tools help laboratories to stay ahead of the curveMass Spectrometry & Spectroscopy Articles- Unlocking the complexity of metabolomics: Pushi...
View all digital editions
Events
Jan 21 2026 Tokyo, Japan
Jan 28 2026 Tokyo, Japan
Jan 29 2026 New Delhi, India
Feb 07 2026 Boston, MA, USA
Asia Pharma Expo/Asia Lab Expo
Feb 12 2026 Dhaka, Bangladesh


















