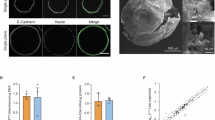
Abstract
Creating in vitro models of diseases of the pancreatic ductal compartment requires a comprehensive understanding of the developmental trajectories of pancreas-specific cell types. Here we report the single-cell characterization of the differentiation of pancreatic duct-like organoids (PDLOs) from human induced pluripotent stem cells (hiPSCs) on a microwell chip that facilitates the uniform aggregation and chemical induction of hiPSC-derived pancreatic progenitors. Using time-resolved single-cell transcriptional profiling and immunofluorescence imaging of the forming PDLOs, we identified differentiation routes from pancreatic progenitors through ductal intermediates to two types of mature duct-like cells and a few non-ductal cell types. PDLO subpopulations expressed either mucins or the cystic fibrosis transmembrane conductance regulator, and resembled human adult duct cells. We also used the chip to uncover ductal markers relevant to pancreatic carcinogenesis, and to establish PDLO co-cultures with stellate cells, which allowed for the study of epithelial–mesenchymal signalling. The PDLO microsystem could be used to establish patient-specific pancreatic duct models.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. Raw data, read counts and the analysed datasets from scRNA-seq can be accessed from the Gene Expression Omnibus repository using the accession code GSE162547. Mass spectrometry data have been deposited on the PRIDE database and can be accessed with the identifier PXD024461.
Code availability
The code for scRNA-seq analysis is available on Zenodo at https://doi.org/10.5281/zenodo.4738625.
References
Lee, M. G., Ohana, E., Park, H. W., Yang, D. & Muallem, S. Molecular mechanism of pancreatic and salivary gland fluid and HCO3 secretion. Physiol. Rev. 92, 39–74 (2012).
Hohwieler, M. et al. Stem cell-derived organoids to model gastrointestinal facets of cystic fibrosis. United European Gastroenterol. J. 5, 609–624 (2017).
Ferreira, R. M. M. et al. Duct- and acinar-derived pancreatic ductal adenocarcinomas show distinct tumor progression and marker expression. Cell Rep. 21, 966–978 (2017).
Bailey, J. M. et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene 35, 4282–4288 (2016).
Lee, A. Y. L. et al. Cell of origin affects tumour development and phenotype in pancreatic ductal adenocarcinoma. Gut 68, 487–498 (2019).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013).
Tuveson, D. & Clevers, H. Cancer modeling meets human organoid technology. Science 364, 952–955 (2019).
Frappart, P. O. et al. Pancreatic cancer-derived organoids—a disease modeling tool to predict drug response. United European Gastroenterol J. 8, 594–606 (2020).
Tiriac, H. et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Disco. 8, 1112–1129 (2018).
Georgakopoulos, N. et al. Long-term expansion, genomic stability and in vivo safety of adult human pancreas organoids. BMC Dev. Biol. 20, 4 (2020).
Boj, S. F. et al. Model organoids provide new research opportunities for ductal pancreatic cancer. Mol. Cell. Oncol. 3, e1014757 (2016).
Hohwieler, M. et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 66, 473–486 (2017).
Huang, L. et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21, 1364–1371 (2015).
Zhou, Q. et al. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell 13, 103–114 (2007).
Villani, V. et al. SOX9+/PTF1A+ cells define the tip progenitor cells of the human fetal pancreas of the second trimester. Stem Cells Transl. Med. 8, 1249–1264 (2019).
Schaffer, A. E., Freude, K. K., Nelson, S. B. & Sander, M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev. Cell 18, 1022–1029 (2010).
Jennings, R. E. et al. Development of the human pancreas from foregut to endocrine commitment. Diabetes 62, 3514–3522 (2013).
Breunig, M. et al. Modelling plasticity and dysplasia of pancreatic ductal organoids derived from human pluripotent stem cells. Cell Stem Cell 28, 1105–1124 (2021).
Huang, L. et al. Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell Stem Cell 28, 1090–1104 (2021).
The Tabula Muris Consortium Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
Enge, M. et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330 (2017).
Baron, M. et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360 (2016).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394 (2016).
Qadir, M. M. F. et al. Single-cell resolution analysis of the human pancreatic ductal progenitor cell niche. Proc. Natl Acad. Sci. USA 117, 10876–10887 (2020).
Segerstolpe, Å. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607 (2016).
Wang, Y. J. et al. Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65, 3028–3038 (2016).
Hwang, Y. S. et al. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc. Natl Acad. Sci. 106, 16978–16983 (2009).
Brandenberg, N. et al. High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat. Biomed. Eng. 4, 864–874 (2020).
Freyer, J. P. Role of necrosis in regulating the growth saturation of multicellular spheroids. Cancer Res. 48, 2432–2439 (1988).
Crisera, C. A. et al. Expression and role of laminin-1 in mouse pancreatic organogenesis. Diabetes 49, 936–944 (2000).
Miner, J. H. & Yurchenco, P. D. Laminin functions in tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 20, 255–284 (2004).
Jiang, F. X., Cram, D. S., DeAizpurua, H. J. & Harrison, L. C. Laminin-1 promotes differentiation of fetal mouse pancreatic beta-cells. Diabetes 48, 722–730 (1999).
Kopp, J. L. et al. Progenitor cell domains in the developing and adult pancreas. Cell Cycle 10, 1921–1927 (2011).
Kopp, J. L. et al. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development 138, 653–665 (2011).
Pan, F. C. & Wright, C. Pancreas organogenesis: from bud to plexus to gland. Dev. Dyn. 240, 530–565 (2011).
Nair, G. & Hebrok, M. Islet formation in mice and men: lessons for the generation of functional insulin-producing β-cells from human pluripotent stem cells. Curr. Opin. Genet. Dev. 32, 171–180 (2015).
Inada, A., Nienaber, C., Fonseca, S. & Bonner-Weir, S. Timing and expression pattern of carbonic anhydrase II in pancreas. Dev. Dyn. 235, 1571–1577 (2006).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Puri, S., Folias, A. E. & Hebrok, M. Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell 16, 18–31 (2015).
George, N. M., Day, C. E., Boerner, B. P., Johnson, R. L. & Sarvetnick, N. E. Hippo signaling regulates pancreas development through inactivation of Yap. Mol. Cell. Biol. 32, 5116–5128 (2012).
Qu, H. et al. Laminin 411 acts as a potent inducer of umbilical cord mesenchymal stem cell differentiation into insulin-producing cells. J. Transl. Med. 12, 135 (2014).
Mamidi, A. et al. Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature 564, 114–118 (2018).
Cirulli, V. et al. Expression and function of αvβ3 and αvβ5 integrins in the developing pancreas: roles in the adhesion and migration of putative endocrine progenitor cells. J. Cell Biol. 150, 1445–1460 (2000).
Erkan, M. et al. StellaTUM: current consensus and discussion on pancreatic stellate cell research. Gut 61, 172–178 (2012).
Uhlén, M. et al. The human secretome. Sci. Signal 12, eaaz0274 (2019).
Uhlen, M. et al. A pathology atlas of the human cancer transcriptome. Science 357, eaan2507 (2017).
Zubair, H. et al. Proteomic analysis of MYB-regulated secretome identifies functional pathways and biomarkers: potential pathobiological and clinical implications. J. Proteome Res. 19, 794–804 (2020).
Bandaru, S. et al. Targeting filamin B induces tumor growth and metastasis via enhanced activity of matrix metalloproteinase-9 and secretion of VEGF-A. Oncogenesis 3, e119 (2014).
Iguchi, Y. et al. Filamin B enhances the invasiveness of cancer cells into 3D collagen matrices. Cell Struct. Funct. 40, 61–67 (2015).
Surcel, A. et al. Targeting mechanoresponsive proteins in pancreatic cancer: 4-hydroxyacetophenone blocks dissemination and invasion by activating MYH14. Cancer Res. 79, 4665–4678 (2019).
Arnold, F. et al. RINT1 regulates SUMOylation and the DNA damage response to preserve cellular homeostasis in pancreatic cancer. Cancer Res. 81, 1758–1774 (2021).
Feld, F. M. et al. GOT1/AST1 expression status as a prognostic biomarker in pancreatic ductal adenocarcinoma. Oncotarget 6, 4516–4526 (2015).
Schmid, S. J. et al. Absence of FLICE-inhibitory protein is a novel independent prognostic marker for very short survival in pancreatic ductal adenocarcinoma. Pancreas 42, 1114–1119 (2013).
Hirsch, F. R. et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol. 21, 3798–3807 (2003).
Kleger, A., Perkhofer, L. & Seufferlein, T. Smarter drugs emerging in pancreatic cancer therapy. Ann. Oncol. 25, 1260–1270 (2014).
Reichert, M., Blume, K., Kleger, A., Hartmann, D. & von Figura, G. Developmental pathways direct pancreatic cancer initiation from its cellular origin. Stem Cells Int. 2016, 9298535 (2016).
Hassid, B. G. et al. Absence of pancreatic intraepithelial neoplasia predicts poor survival after resection of pancreatic cancer. Pancreas 43, 1073–1077 (2014).
Bremer, S. C. B. et al. Enhancer of zeste homolog 2 in colorectal cancer development and progression. Digestion 102, 227–235 (2021).
Li, J. et al. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. eLife 7, e37184 (2018).
Driehuis, E. et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl Acad. Sci. USA 116, 26580–26590 (2019).
Co, J. Y. et al. Controlling epithelial polarity: a human enteroid model for host–pathogen interactions. Cell Rep. 26, 2509–2520.e2504 (2019).
Veres, A. et al. Charting cellular identity during human in vitro β-cell differentiation. Nature 569, 368–373 (2019).
Krentz, N. A. J. et al. Single-cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors. Stem Cell Rep. 11, 1551–1564 (2018).
Petersen, M. B. K. et al. Single-cell gene expression analysis of a human ESC model of pancreatic endocrine development reveals different paths to β-cell differentiation. Stem Cell Rep. 9, 1246–1261 (2017).
Wang, J., Yuan, R., Zhu, X. & Ao, P. Adaptive landscape shaped by core endogenous network coordinates complex early progenitor fate commitments in embryonic pancreas. Sci. Rep. 10, 1112 (2020).
Chen, C. et al. Evidence of a developmental origin for β-cell heterogeneity using a dual lineage-tracing technology. Development 146, dev164913 (2019).
Larsen, H. L. et al. Stochastic priming and spatial cues orchestrate heterogeneous clonal contribution to mouse pancreas organogenesis. Nat. Commun. 8, 605 (2017).
Gitlin, L., Schulze, P. & Belder, D. Rapid replication of master structures by double casting with PDMS. Lab Chip 9, 3000–3002 (2009).
Nostro, M. C. et al. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Rep. 4, 591–604 (2015).
Mohammad Rezazadeh, F. et al. Fast free of acrylamide clearing tissue (FACT) for clearing, immunolabelling and three-dimensional imaging of partridge tissues. Microsc. Res. Tech. 81, 1374–1382 (2018).
John, T., Liu, G. & Tsao, M. S. Overview of molecular testing in non-small-cell lung cancer: mutational analysis, gene copy number, protein expression and other biomarkers of EGFR for the prediction of response to tyrosine kinase inhibitors. Oncogene 28, S14–S23 (2009).
Jesnowski, R. et al. Immortalization of pancreatic stellate cells as an in vitro model of pancreatic fibrosis: deactivation is induced by matrigel and N-acetylcysteine. Lab Invest. 85, 1276–1291 (2005).
Livnat-Levanon, N. et al. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 7, 1371–1380 (2014).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–w198 (2019).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Chen, E. Y. et al. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics 28, 105–111 (2012).
Clarke, D. J. B. et al. eXpression2Kinases (X2K) Web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res. 46, W171–W179 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Guillaumet-Adkins, A. et al. Single-cell transcriptome conservation in cryopreserved cells and tissues. Genome Biol. 18, 45 (2017).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Luecken, M. D. & Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech: Theory Exp. 2008, P10008 (2008).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Bastidas-Ponce, A. et al. Comprehensive single cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development 146, dev173849 (2019).
Tenenbaum, D. KEGGREST: client-side REST access to KEGG. R package version 1.24.1 (2019).
Acknowledgements
This work is supported by the Helmholtz Pioneer Campus, the German Ministry of Education and Research (INDIMED-PancChip), the Baden-Württemberg Stiftung (project ExPoChip), ERC (Consolidator Grant Number 772646), Deutsche Forschungsgemeinschaft (DFG) Sachbeihilfe KL 2544/7-1, KL 2544/1-2, KL 2544/5-1, GRK 2254/1 and 2, Heisenberg-Programm KL 2544/6-1, German Cancer Aid (Grant 111879), Else Kröner-Fresenius-Stiftung (supporting A.K. with an Excellence grant and M.H. with a First Application grant) and Bausteinprogramm of Ulm University (granted to M.H.). We thank NK-Optik for instrumental support. We thank M. Löhr (Karolinska Institute) for providing the human pancreatic stellate cells and T. Walzthöni for bioinformatics support provided at the Bioinformatics Core Facility, Institute of Computational Biology, Helmholtz Zentrum München.
Author information
Authors and Affiliations
Contributions
S.W., M.B., M.H., A.K. and M. Meier designed the study. S.W. designed and produced the microwell chips, based on work from M. Moussus. S.W., M.B., J.M. and M.H. executed the biological experiments. S.W., M.B., M.H. and T.G. did the imaging and image analysis. S.W., M.S. and H.L. performed the scRNA-seq processing and S.W. did the analysis. C.v.T. and S.M.H. did the mass spectrometric measurements and data processing. S.E.W. and P.M. stained and evaluated the FLNB patient cohort. L.S. and T.S. took the serum samples of the PDAC patient cohort and M.H. performed the ELISA. S.W., M.B. and M. Meier analysed the MS/MS and FLNB screening results. M.H., A.K. and M. Meier received the funding and supervised the study. The manuscript was written by S.W., M.B., M.H., A.K. and M. Meier. All authors corrected and approved the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biomedical Engineering thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures.
Supplementary Dataset 1
Top 300 DEGs for the kinetic clusters in Fig. 4c.
Supplementary Dataset 2
Dynamical genes for Fig. 6f.
Supplementary Dataset 3
Top 300 DEGs for the Louvain clusters in Fig. 7a.
Supplementary Dataset 4
PDLO secretome and proteome.
Supplementary Dataset 5
Results of the ELISA FLNB screening corresponding to Fig. 8h.
Supplementary Dataset 6
Antibodies used for IHC/IF-p, ICC/IF and FC staining.
Supplementary Video 1
Live-cell imaging during PDLO differentiation from day 24 until day 31.
Rights and permissions
About this article
Cite this article
Wiedenmann, S., Breunig, M., Merkle, J. et al. Single-cell-resolved differentiation of human induced pluripotent stem cells into pancreatic duct-like organoids on a microwell chip. Nat Biomed Eng 5, 897–913 (2021). https://doi.org/10.1038/s41551-021-00757-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00757-2
This article is cited by
-
Scalable production of uniform and mature organoids in a 3D geometrically-engineered permeable membrane
Nature Communications (2024)
-
A pancreatic cancer organoid-in-matrix platform shows distinct sensitivities to T cell killing
Scientific Reports (2024)
-
Exploring human pancreatic organoid modelling through single-cell RNA sequencing analysis
Communications Biology (2024)
-
Translational Utility of Organoid Models for Biomedical Research on Gastrointestinal Diseases
Stem Cell Reviews and Reports (2024)
-
TBX3 is dynamically expressed in pancreatic organogenesis and fine-tunes regeneration
BMC Biology (2023)