This is a download from Nature Research, part of Springer Nature.
Geoff Marsh
Hello. Welcome back to the podcast where we celebrate the winner of the Eppendorf Award for Young European Investigators. This year is a special year, marking the 25th anniversary of the awards which have been run in partnership with Nature since 1995. Usually, my introduction is recorded in the jam-packed foyer of the EMBL’s Advanced Training Centre in Heidelberg, Germany. But as with virtually all conferences this year, COVID-19 has put a stop to the proceedings. So instead, I've turned to video conferencing to meet this jubilee year winner, Professor Randall Platt. This isn't exactly how I expected to meet you. You're in your living room it looks like. I'm in my living room. We’re having to do this remotely, but it's nice to meet you.
Randall Platt
It's great to meet you too. I’m calling from Basel, Switzerland.
Geoff Marsh
So, first of all, just tell me who you are.
Randall Platt
So, my name is Randall Platt. I go by Randy. I'm currently an assistant professor. I have a dual appointment. One is at the ETH Zurich in the Department of Biosystems Science and Engineering, and the other one is at the University of Basel and the Department of Chemistry, and I've been in Basel for just about four years.
Geoff Marsh
Okay, well, first and foremost, congratulations on winning this highly prestigious research prize and on this iconic anniversary year, 25th anniversary year. It's a shame that we're doing this remotely, but you must feel chuffed.
Randall Platt
It is indeed a tremendous honour, and the fact that it's virtual, yeah, it's unfortunate to say the least, especially the all-star cast of speakers that were planned for this jubilee. So, yeah, we're missing out, but we'll come back next year and it will be okay.
Geoff Marsh
The technology that you've been awarded the prize for primarily, I believe, is a type of molecular recorder called Record-seq. What is a molecular recorder?
Randall Platt
Yeah, so, a molecular recorder is an emerging class of molecular technology. It's basically a collection of genes that when put inside of cells endow them with a synthetic form of memory. So, a molecular recorder could, say, take a biological signal of the world – a chemical, a molecule, a transcription factor – transduce that into, say, DNA changes in a DNA storage medium and, through sequencing, we can kind of reconstruct that synthetic cellular memory and identify what the biological signal is.
Geoff Marsh
So, you've adopted this system from microbes. What is that system doing for microbes in nature?
Randall Platt
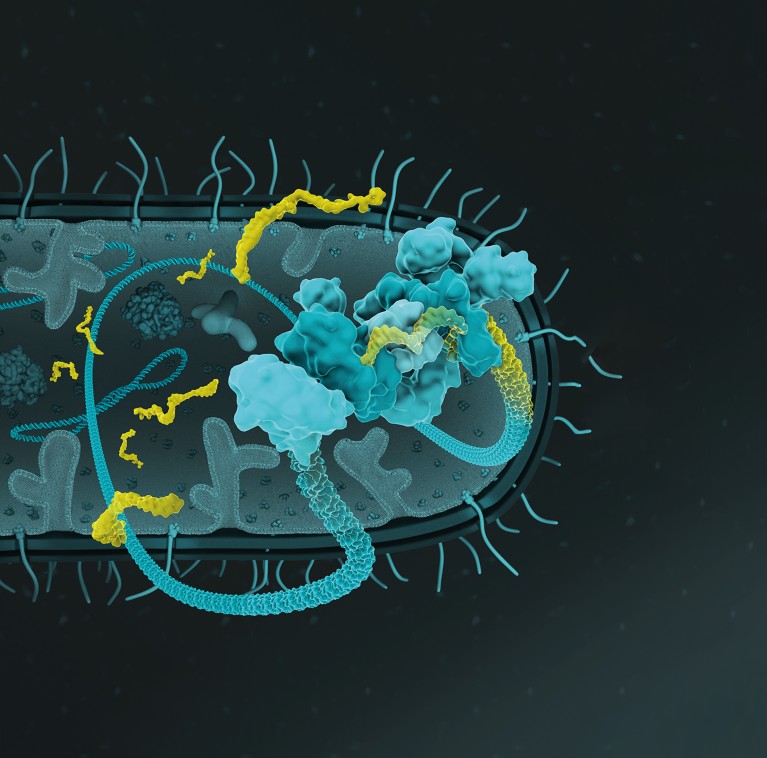
Yeah, so CRISPR formerly is considered what is called an adaptive immune system. This is what allows your body to remember the viruses that you're infected by and provide you with resistance with future infections. You have a bacterial cell, it's infected with a virus, the virus injects its genetic material, DNA or RNA, and then the CRISPR system has an army of proteins that execute different functions. So, some of these proteins will basically take a piece of that virus and insert it in its own genome, where it serves as a molecular memory of that invader. And then at a later point in time, the CRISPR system can recall that memory in the form of a small piece of RNA called a guide RNA which then guides CRISPR-associated enzymes back to viral invaders, destroys them by cutting them and thus provides the bacteria with immunity. And so, my lab, as well as the labs of others, have been hijacking these components for interesting applications in biology and medicine. The most popular application is those that rely on one CRISPR enzyme called Cas9, and people use this as molecular scissors to modify genes to, say, correct a genetic mutation and cure a disease. But CRISPR is this toolbox of proteins and so our advance specifically focuses on that first step of CRISPR, the recording part where the virus’ genome is inserted into the bacterial genome. That process is recording, right, and so we're leveraging that recording process specifically to endow other cells that we want to be able to do the same thing, which is basically record RNA which we can use to predict gene expression over time.
Geoff Marsh
How do you then decode that information into something useful? Is the information stored in a comprehendible format?
Randall Platt
So, this Record-seq process, so if we zoom inside of the cells and look at the molecular mechanism of how these records are formed, there are these CRISPR proteins inside of cells. They form a complex, so this big multi-protein complex, and they essentially grab RNAs proportional to their abundance, and they take a little snippet of them, about 40 nucleotides, and then they integrate that within what is called a CRISPR array – this is just a DNA sequence of repetitive records. And this recording happens in a very interesting way. The records are formed iteratively and sequentially such that after multiple recordings you can have essentially a fossil record where you have these records perfectly aligned topologically such that you know the order of events. So, we're not only able to record whether a gene was expressed, but we're able to tell when it was expressed.
Geoff Marsh
That's so handy that it works that way.
Randall Platt
It's very convenient. Where we're specifically applying it now and where I think it can have immediate value is in the area of living diagnostics. So, for example, men over a certain age routinely gets colonoscopies to check for colorectal cancer, right? This is a microscope inserted up one side of your body and with a camera looking for small masses. Now, imagine an alternative example where a physician could give you a yogurt and this contains bacteria. You could do this from your home. You could send in your sample, and the same cells could tell the doctor or clinicians whether you have an early stage polyp or you have inflammatory bowel disease. So, our goal is to evaluate to what extent these recording cells can be applied in this context of recording disease pathology via the gastrointestinal tract.
Geoff Marsh
So, in order to get these microbial sentinel cells to keep a record of something, they need to express a gene in response to it, so presumably a challenge for you is engineering these bacterial cells that respond to specific molecules.
Randall Platt
You landed on the very interesting crux of the problem – we need to infer human environments from bacterial gene expression. So, our technology, which we call Record-seq, can capture gene expression at the entire transcriptome scale. So, we can get an observation of most genes and E. coli, for now. And so, with those records of E. coli gene expression, we need to be able to predict what the outside environment looks like. In some cases, this can be quite trivial, so we've already started performing these experiments in mice under different diets, under different disease settings, and for the most part, it results in a fairly predictable response. So, for example, if you induce colitis, which is inflammation of the large intestine, you would expect a specific environment – a reactive oxygen species that would, in turn, act on the E. coli in a specific way. They basically upregulate the expression of protective factors and if we look in our transcriptional records, as we call them, we can indeed find this stress response. We're quite fortunate in that evolution endowed bacteria with a plethora of sensors for a range of molecules, right? These are single-celled organisms that need to survive in tremendously diverse environments. Compared to us, bacteria are masters of different environments. E. coli, for example, can live in the soil, they can live in a culture tube in your lab, and they can also live inside your intestine. And so, with this natural arsenal of sensors, it gives us a lot to work with. We could also imagine not using E. coli but other bacteria with a different arsenal of sensors. So, we think this is where these open areas for future developments lie.
Geoff Marsh
I imagine clinicians and patients would be excited by a technology like Record-seq, as you say, because it could, you know, negate the need for invasive procedures. How close do you believe Record-seq is to the clinic?
Randall Platt
With any radically new technology, the timeline between development to implementation in a person is a long one, so this isn't happening tomorrow. This might not even be happening in this decade. We don't really know. It depends on a number of factors based on, say, how well the system works, how specific it is for a given disorder and also, there is an unclear regulatory path in terms of using genetically modified bacteria inside of people. And so, this is a big unknown, really, so we still have a lot of developments to do in the lab. The experiments in mice are incredibly promising. But there's a number of engineering and quality control clinical development steps that we need to do.
Geoff Marsh
I imagine you can envisage multiple applications for a technology like Record-seq.
Randall Platt
So, while we are choosing to apply it in the case of living diagnostics via the gut, it's true that this can be applied elsewhere. Bacterial cells and microbes cover every surface of this planet at high densities, some more than others, but they're everywhere. If you embed recording devices inside of these cells, you could seemingly create sentinels of any environment. We're using the body, but you could potentially put this in soil bacteria, you could put it in vector in the ocean or the air to monitor, say, environmental contaminants. Another area that we're working on is also adapting the system to eukaryotic cells so we can start to interrogate these cell transitions in the context of human development and disease.
Geoff Marsh
Do you reach an upper limit in terms of the storage capacity of a microbe? They're tiny.
Randall Platt
So, one of the limitations of the technology now is the efficiency of recording. I’ve painted the picture where a cell acquires multiple records. This is indeed true, but over a population, which is required for the Record-seq technology, the efficiency is, frankly, not great. So, as for storage capacity, we're nowhere near the fundamental limit. But if we use nature as a guide, we can look throughout different microbes and see how large these CRISPR arrays can get, and I think this defines a theoretical bound, and there's one bacterial species that harbours around 300. So, if you imagine a population of a million bacteria each containing 300 records, and given that the records are arranged very precisely in this CRISPR array from old to new, you can start to use this population reconstruction to potentially reconstruct gene expression transcriptome scale throughout time.
Geoff Marsh
What's next then for you and your lab?
Randall Platt
So, in the future, we have ambitions for continuing to develop Record-seq in the context of human health. But outside of that, my lab also develops other molecular technologies with different goals in mind, and these areas are more in line with genetic disorders, understanding how they arise, and coming up with genomic-based treatments for their cure. And so, we've created a few different technologies based on CRISPR enzymes that allow us to massively change genomes and transcriptomes toward correcting complex genetic disorders. That's one example. We're also developing other techniques in functional genomics to increase the scale at which we perturb and understand gene function.
Geoff Marsh
I have to say it sounds like an exciting time to be a team member in in your lab.
Randall Platt
I don't know, you would definitely have to ask the team. But with that, I really would like to acknowledge the entire team. Over the first four years here, I've been able to recruit an exceptional group of people and I'm thrilled to have each and every one of them, so huge thanks to all of them. And since we're acknowledging those that had an impact on me and my work, I'd also like to acknowledge my mentors and my collaborators, as well as my family for their support. So, thank you for that question.
Geoff Marsh
Well, thank you for that wonderful discussion, Randy. Before I end the podcast, I wanted to hear from one of the jury members who’d chosen Randy for this year's award. Professor Laura Machesky was kind enough to oblige. She started off with her full introduction.
Laura Machesky
So, I'm Professor Laura Machesky and I work at the Beatson Institute for Cancer Research at the University of Glasgow.
Geoff Marsh
We usually meet at the EMBL. It's a shame that we're not there this year.
Laura Machesky
I know, that's right. It's such a nice place to go for a reception and to hear the talks from the prize winner, so we really missed it this year because Randy's been a great candidate and it would have been very exciting to hear him live talking about his work. But it still doesn't take away from the great accomplishment that he's that he's done.
Geoff Marsh
What was your experience of being a jury member again?
Laura Machesky
It's always a fun and exciting experience being a jury member because we have so many applications and nominations of excellent, outstanding, young investigators and, for us, it's a chance to find out what kind of exciting science and technology is having happening around Europe and to really see profiles of some of the best and most talented researchers in European science.
Geoff Marsh
Could you just give me a sense of the rapid rise of CRISPR technology more broadly?
Laura Machesky
I remember working in the lab when we were excited about the idea of siRNA and being able to silence genes that we were working on rather than having to rely on things like chemical tools or dominant negatives only. And so, CRISPR was super exciting, you know, even coming after siRNA and really, it's only been the last, I don't know, 5-10 years when that technology's been available widely in laboratories for scientists to be able to kind of use precision methods to knock out or modify genes that they're interested in and enabled us to study genes in a much more clean and clear way. And then the advantages and medical possibilities arising from CRISPR has been amazing. I mean, it hasn't been yet realised that CRISPR, to my knowledge, is used for medical therapy but certainly there are so many hopeful applications of CRISPR to the medical field that, if we could use that precision to address genetic diseases, it has amazing potential.
Geoff Marsh
CRISPR has kind of taken the scientific world by storm and it's amazing that then Randy came along, saw this group of proteins and dreamt up this completely novel use for them.
Laura Machesky
It is amazing, and I think Randy seems to be extremely creative in his approach to science, so he doesn't just do the next step, and I think he was creative as an undergraduate. He went off and did things that his supervisor was not necessarily in favour of and convinced his supervisor that these different aspects of his project were worth pursuing. And I think he's always had a little bit of that, it seems like, fearlessness and creativity to go beyond what the obvious next step is and actually to do something new and see something new in what he's doing that really stands him out from the crowd.
Geoff Marsh
He now heads his own lab, doesn't he, at ETH Zurich. How often do you see researchers progressing at this speed?
Laura Machesky
He has progressed very fast. So, he went right from his PhD, only did a short postdoc – less than a year – where he was co-advised by Feng Zhang and Paola Arlotta at Harvard, and this was a collaboration that he initiated, where he's applying genome editing in utero in mice to modify mice before they're born. And this is a project that he's still actually working on. But that was a seven-month transition to his independent position, and so it's very rare for a person to go right from a PhD to being a lab supervisor. So, ETH Zurich in Basel really had the insight to know that Randy was one of those creative people that was ready to be independent and ready to start his own group. It's clear from his success in his early years of running his lab there that they made a great choice.
Geoff Marsh
You're a cancer researcher. Whilst we've been talking about the importance of basic research and what that can lead to, Record-seq holds promise for the clinic, doesn't it? As a cancer researcher yourself, how excited are you by this new technology?
Laura Machesky
I think it's very exciting. I mean, I think the idea that Record-seq could be used to develop less invasive tests for reporting on the human experience during disease or life events is amazing. And so, it's still obviously under development but the idea that you could make a yogurt drink which contains these microbes that can record events in your body, and you could drink your yogurt and then have the bacteria sequenced to know what they're experiencing, so are they in a cancer environment or are they in an infection environment in your body, without you having to have invasive testing, it's very attractive. And whether it will be realised and how long it will take, time will tell, but it's very intriguing, and it also shows the way he's thinking outside of the box and developing these ideas.
Geoff Marsh
Okay, well, thank you for chatting to me over Zoom from your office, and I hope next time we meet we'll be with champagne at the EMBL.
Laura Machesky
That would be fantastic. I hope so.
Geoff Marsh
As you know, this year’s event has been postponed until 2021, but then there'll be two years’ worth of winners to celebrate, as well as many other winners from the previous 25 years. We hope to see you there. This has been a special podcast from Nature Research, part of Springer Nature. I'm Geoff Marsh. Thank you for listening.